SCRIPTS¶
Introduction¶
The are some typical scenarios of working with input and output data of KiSSAM. The software package includes a variety of scripts for pre- and postprocessing.
The scripts are used through the Linux command line. To use a script, the following patters should be written in the bash terminal
<script name> <option(s)> <arguments(s)> [<optional argument(s)>]
The <script name> includes the path to the KiSSAM instalation folder. In the instructions below, we use angle brackets to indicate arguments that should be filled in, and square brackets to indicate optional arguments.
Run any script without options for the brief explanation of its options.
Running Scripts with docker¶
If you have obtained the docker version of KiSSAM, the scripts are run in the docker as well. Use the command that you have run KiSSAM with, and add the script command at the end of it. For example
docker run --gpus all -e USER_ID=$(id -u) -e GROUP_ID=$(id -g) -e INPUT_JSON_FILE=input.json --rm -v .:/calc -it kissam "<script name> <option(s)> <arguments(s)> [<optional argument(s)>]"
Simulation Preparation Scripts¶
Powder Sphere Rasterization¶
rasterizeSpheres.x rasterizes spheres from text list into the VDB format. VDB file contains volumetric data: the filling fraction of material on a fine mesh. Usage:
/opt/KiSSAM/scripts/rasterizeSpheres.x <VoxelSize> <file>
<file> (for example, spheres.dat) should be a text file with the four columns: \(x\), \(y\), \(z\) coordinates of the sphere position and radius (in micrometers).
<VoxelSize> is the mesh step size in micrometers.
27.4911 27.4917 27.4911 27.5
44.7133 79.1247 63.0167 37.5
87.07 32.4956 32.4953 32.5
166.9 34.6008 27.4979 27.5
128.672 75.4377 27.498 27.5
...
After that the file init_spheres.vdb will be created.
To use it in the simulation, specify its name and file path in the JSON field Powder.initSpheresVDBfile.
Make sure that the mesh step size in the KiSSAM simulation configuration (default.json, or the input JSON) in NumericalParams.dr is equal to <VoxelSize> specified here.
Note that NumericalParams.dr is specified in meters, while <VoxelSize> is specified in micrometers.
G-code Converter¶
The helper script cnc-to-XY.py can prepare the source file for the scanning path setting XYpositions from the cnc files in g-code.
/opt/KiSSAM/scripts/cnc-to-XY.py <filename> --config <input.json>
Here, <filename> is the input file in g-code, and input.json is the simulation configuration file.
In the JSON file, the following additional setting is required for the G-code converter script:
"G-code-converter": {
"idleSpeed": 7000,
"workSpeed": 850,
"offset": [25,25]
}
Please note that there are many g-code file formats, and some may be not supported in the current version.
If the script is unable to process your files, please contact the KiSSAM support or prepare the XYpositions file manually.
Post Processing Scripts¶
Visualization helpers¶
Combine images¶
During the simulation, images are written into the rendered3D directory, as well as several subdirectories in the cross-sections/ directory. To combine the images that correspond to one time instant in one image, use the combine_images.sh script.
/opt/KiSSAM/scripts/combine_images.sh <simulation_directory> [--overlay] [--dr <dxstep>]
The arguments in the square brackets are optional.
- -dr <dxstep>¶
Specify the pixel size in micrometers. Should be equal to the NumericalParams.dr used in the simulation. If the option is added, the lines at Y-section image at correct places of X-sections are drawn over the image.
- --overlay¶
If this option is used, the combined image has not only all the images on the current time, but also the images on the previous times. The current image always contains only the melt pool region. To see the previously solidified images as well, turn on
--overlay.
Example. In the directory, where the simulation was started, execute the command:
/opt/KiSSAM/scripts/combine_images.sh output --overlay
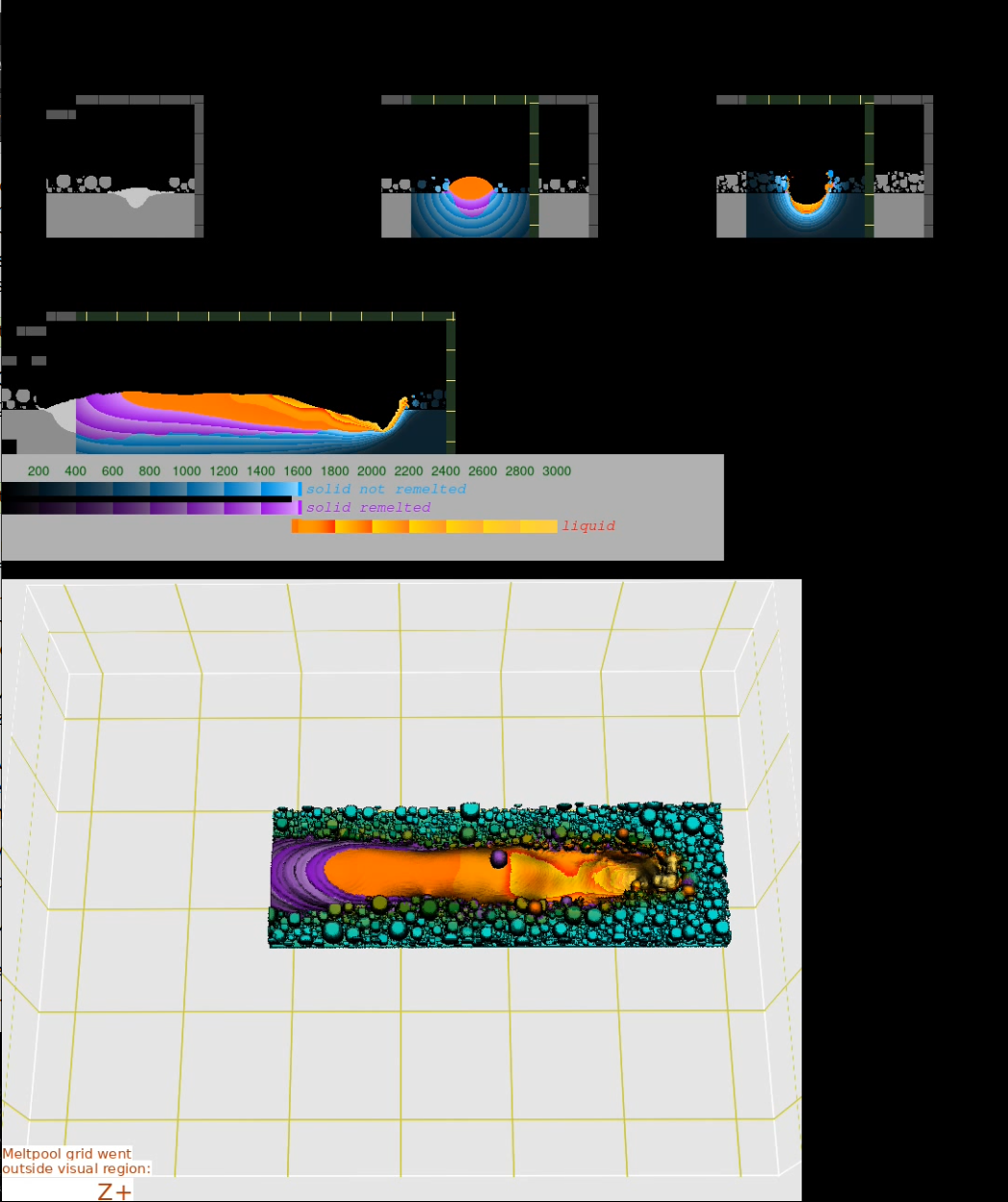
Fig. 6 Result of the script with the overlay option.¶
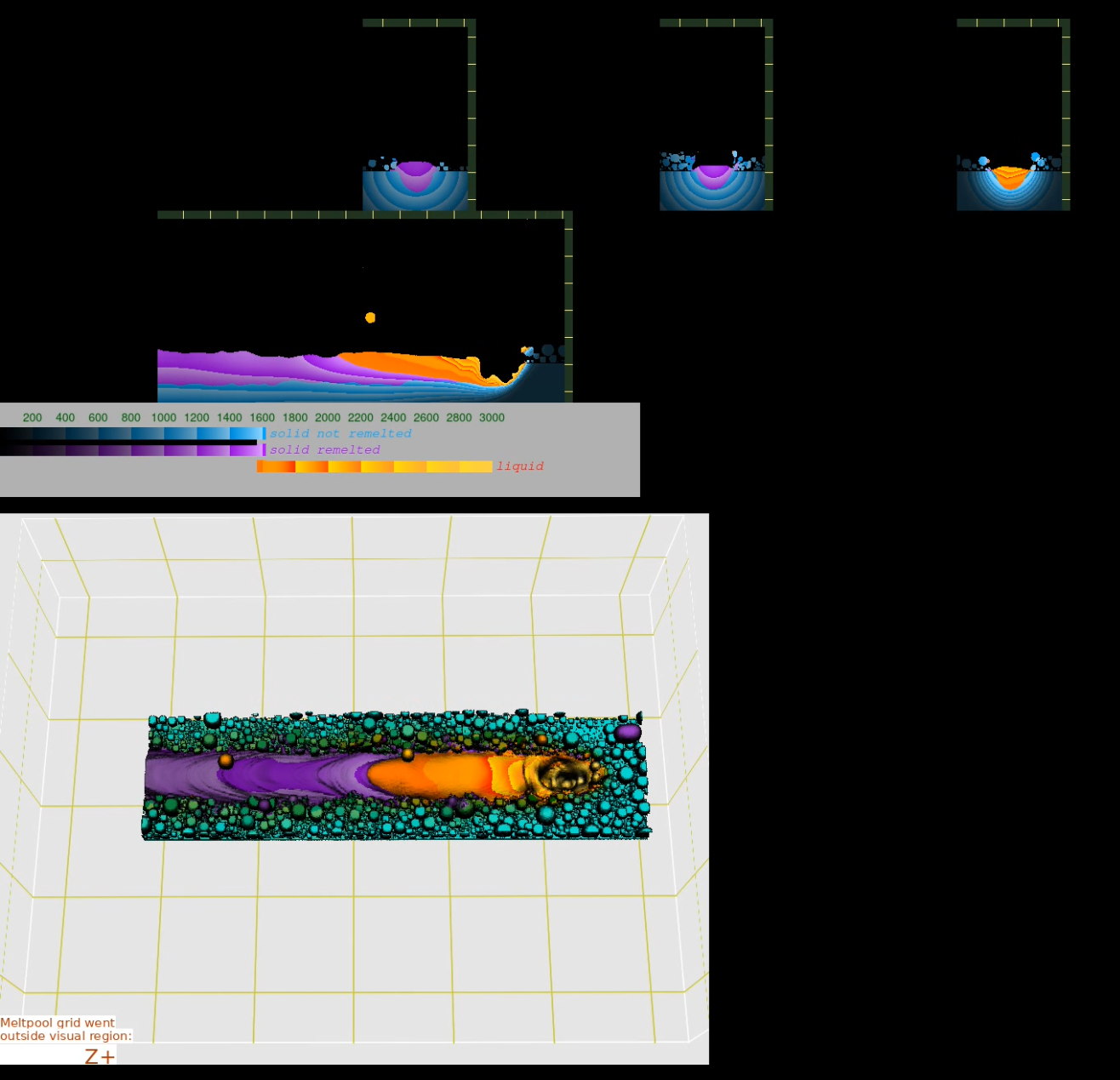
Fig. 7 Result of the script without the overlay option.¶
The result is written in the output/combinedIMG folder. At the same time, the animation of the images is generated and can be found in the output/combinedIMG/video.wmv file. Note that picture files in the cross-sections/ and rendered3D/ directories must have correct file names with cycle iteration ratio of 1000.
Pad images¶
Since the meltpool grid adapts to the size of the meltpool, the images in different time steps have different sizes. This is inconvenient for many purposes, such as animations and slideshows. Moreover, the cross sections in the planes with no melted material are often represented with empty images. The pad_images.sh script creates images with equal width and length and easily to batch them in video later. This script is used by the combine_images.sh script.
Generate Cross Section Images¶
The script drawCrossSection.x generates cross-sections pictures from the geometry.vdb file.
Usage:
./drawCrossSection.x geometry.vdb [ section-pic.pam | section-pic.png ] --sec x|y|z coordinate | --box Xmin,Ymin,Zmin Xmax,Ymax,Zmax
[ --autoclip ]
[ --grains ]
[ --ipf AXIS ]
[ --hl_bound_grains ]
[ --color-grain-size]
[ --color-grain-elong]
[ --grain-stat-2D]
[ --background COLOR ]
[ --substrate COLOR ]
[ --powder COLOR ]
[ --remelted COLOR ]
[ --smooth ]
[ --comp_limit lim1,lim2 ]
[ --comp_color COLOR ]
Output image file should be specified either with .pam or .png extenstion.
The section can be set either with the --sec option or with the --box option.
All the values must be specified in millimeters units.
If the --sec option is used, one of x, y or z symbols should be chosen and the coordinate should be given.
The cross section is taken on that coordinate
If the --box option is used,
one, and only one of the following conditions must be fullfied: xmin == xmax, or ymin == ymax or zmin == zmax.
If xmin == xmax, the output image <output-picture.ppm> is a section of the geometry where
ymin\(<y<\) ymax,
zmin\(<z<\) zmax.
Cross sections in the other two perpendicular planes are made if zmin == zmax or ymin == ymax.
- --autoclip¶
Crop the area where there is no remelted geometry
- --grains¶
Color the grains if the grain growth model for simulation of microstructure was turned on in the simulation
- --ipf AXIS¶
The color of the grains is according to the inverse pole figure (IPF). AXIS can be x, y, or z
Sample usage to get IPF at the \(y=1\) mm plane:
./drawCrossSection.x geometry.vdb picture.png --sec y 1 --grains --ipf z
- --hl_bound_grains¶
Highlight the grain boundaries with solid lines
- --color-grain-size¶
The grains color depends on their 3D volume
- --color-grain-elong¶
The grains color depend on their 3D elongation
- --grain-stat-2D¶
Add this option to make the previous two statistics depend not on the 3D shape of the grains, but on the 2D shape ass seen in the cross section
- --background¶
After this option, specify the color of the image background in the HEX format. Default is 000000FF
- --substrate¶
Color of the substrate in the initial geometry. Default is 7F99B2FF
- --powder¶
Color of the powder. Default is 7F99B2FF
- --remelted¶
Color of the melted material. Default is 33E5FFFF
- --smooth¶
Enable anti-aliasing for the surface
- --comp_limit¶
For multicomponent evaporation simulation, set the limits of the first component for visualization
- --comp_color¶
For multicomponent evaporation simulation, set the color of the component. The color is added to the color of the substrate or powder.
Colors are in the RGBA format and can be transparent.
Here is an example how the --box option is used:
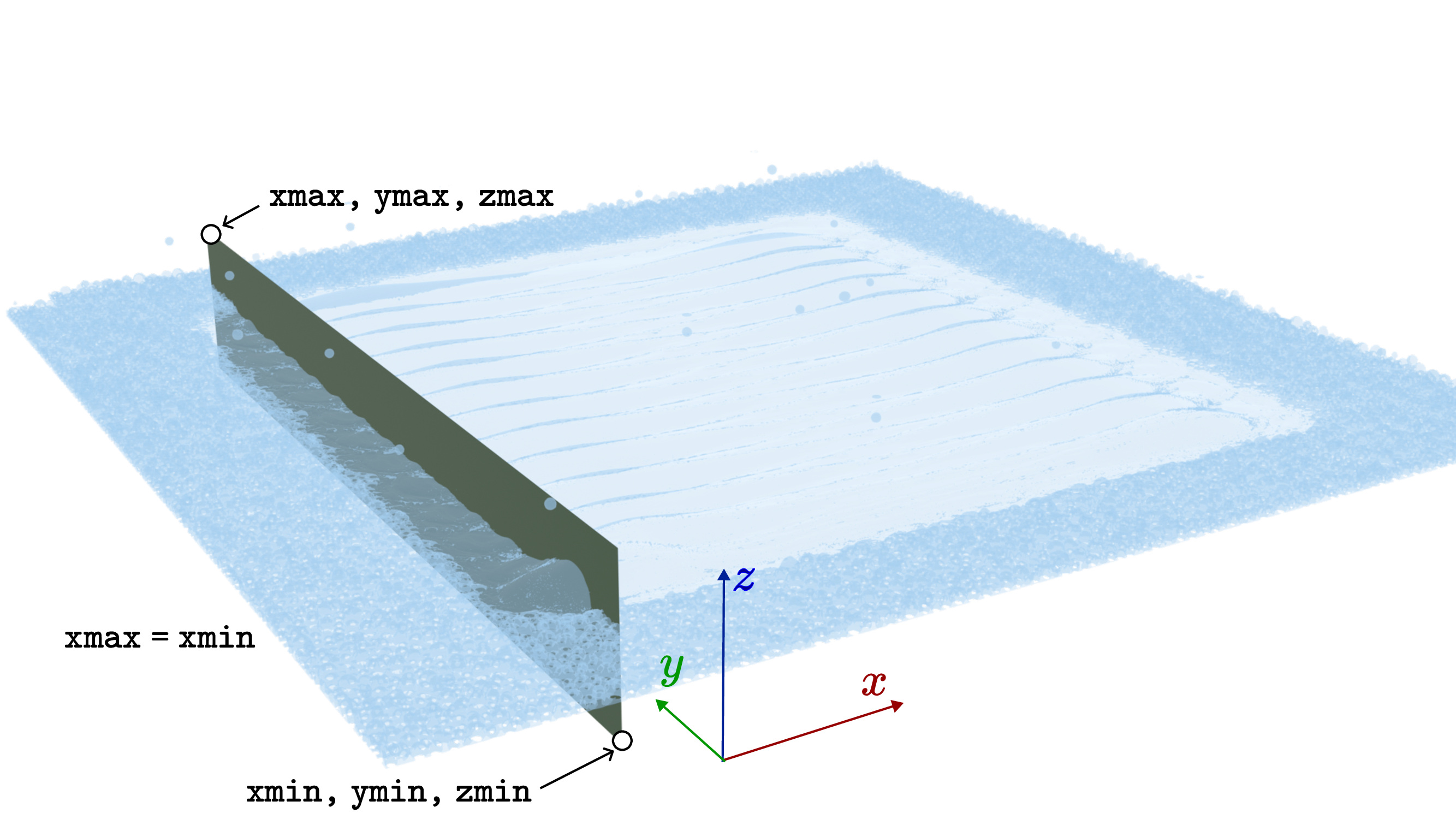
Fig. 8 Illustration of the box parameters.¶

Fig. 9 Cross section result from the previous figure¶
Here is an example. Let us plot cross section for the simulation which was run with the following data in the input JSON file:
"sizes": {
"FullXapprox": 0.008,
"FullYapprox": 0.008,
"FullZapprox": 0.005,
"substrate" : 0.00456
},
"ScanStrategy": {
"Type": "SingleTrack",
"SingleTrack": {
"length" : 5e-3,
"startPosX": 1.0e-3,
"startPosY": 2.5e-3
},
To plot the cross section along the track, we have to specify the \(y=2.5\) mm plane. Thus, ymin = 2.5 = ymax. The cross section box has be around the substrate: zmin < 4.56 mm < zmax. The box has to include the length of the track:xmin < 1 mm < 1+5 mm < xmax. We run the command with
/opt/KiSSAM/scripts/drawCrossSection.x drop/geometry.vdb longCrossSection.png --box 0,2.5,4.4 7,2.5,5
And get the result

Here is an example with specified colors:
/opt/KiSSAM/scripts/drawCrossSection.x drop/geometry.vdb longCrossSection.png --box 0,2.5,4.4 7,2.5,5 --background 00000000 --substrate 7F6D94FF --powder 295569FF --remelted CD86F2FF

Geometry Post-Processing¶
Activate Grids¶
Some of the output VDB files contain grids with all the data, but the cells are not activated, and can not be visualized. In the output geometry VDB file, only the cells that were in the fluid simulation domain are activated, and some of the stationery geometry will not be visualizaed. Use the activateGrids.x script to activate the grids.
/opt/KiSSAM/scripts/activateGrids.x <geometry-input.vdb> <geometry-output.vdb> [--crop <Xmin,Ymin,Zmin Xmax,Ymax,Zmax>]
Rendering of the file with activated geometry can take longer time.
- --crop¶
The option
--cropcan be used to decrease file size. With it, only the box specified by two corners Xmin < \(x\) < Xmax, Ymin < \(y\) < Ymax, Zmin < \(z\) < Zmax (in mm) is written to the output file.
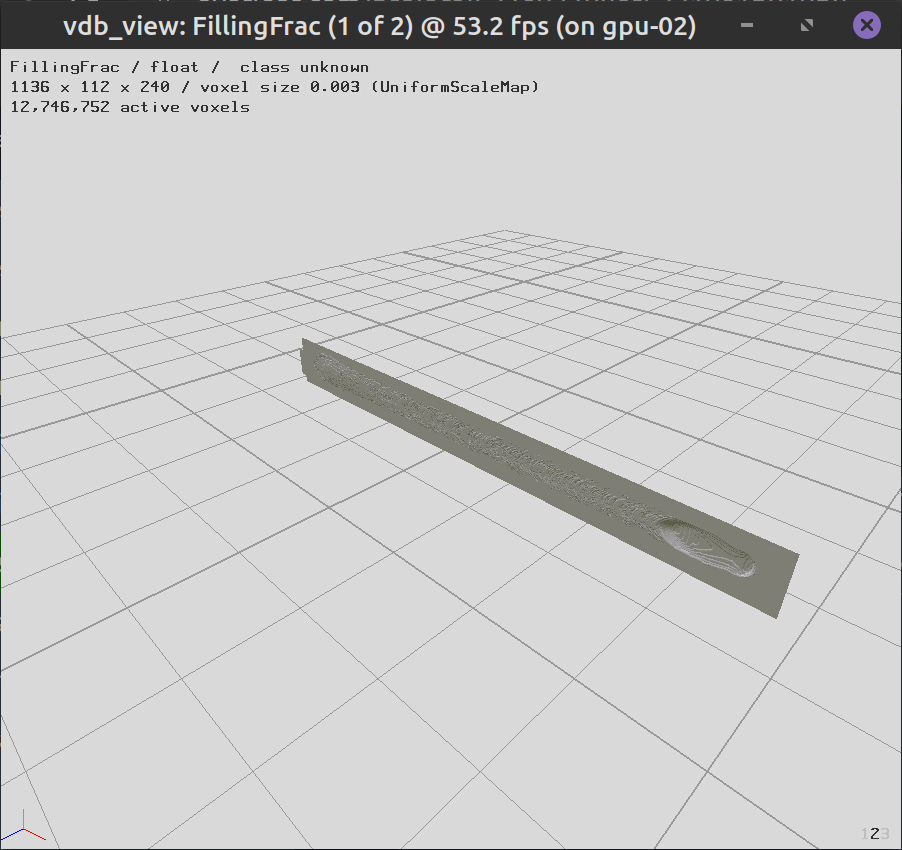
Fig. 10 Before¶
Fig. 11 After¶
Remove Isolated Geometry¶
The script removeIsolated.x removes isolated spatters from geometry.vdb file which are not connected with resulted solidified surface. Usage:
/opt/KiSSAM/scripts/removeIsolated.x geometry.vdb sim_geometry.vdb

Fig. 12 Before (geometry.vdb)¶
Fig. 13 After (sim_geometry.vdb)¶
Remove Powder¶
To study a finished sample after the multilayer simulation is complete, run the removePowder.x script.
/opt/KiSSAM/scripts/removePowder.x
Merge Layers¶
In a multi-layer simulation, the latest geometry.vdb file treats only the last layer as melted and solidified material, and all previous layers as initial substrate.
To find all melted and solidified material across several layers in one VDB file, use the mergeLayers script.
Usage:
./mergeLayers.x <vdb_file1.vdb> <vdb_file2.vdb> <vdb_file3.vdb> ... [--remelted input_files*.vbd] --out <output_file.vdb> [--debug]
- --remelted¶
If the
--remeltedoption is specified, in this case the remelted regions will be picked up only from the selected files.
- --secern¶
If the
--secernoption is specified, the remelted part of different vdb files will be marked with a different value in the ParticleIds grid in the resulting VDB file.
Convert into Triangulated 3D Surface¶
The script vdb2stl.x creates an STL-file of the surface geometry (based on the FillingFrac field of the VDB file). Usage:
/opt/KiSSAM/scripts/vdb2stl.x <vdb_file> <stl_file> [--only-active]
The output file size is often very large.
- --only-active¶
To suppress non-active regions run with the
--only-activeoption.
To reduce the file size by simplifying geometry, use the decimate_stl.py script.
Simplify Surface¶
The script decimate_stl.py decimates an STL file (reduces the total number of polygons). This is used to prepare a cost-efficient input for the powder generation software.
python3 /opt/KiSSAM/scripts/decimate_stl.py surface.stl
Cleanup Surface¶
Process STL file to remove all internal triangles.
/opt/KiSSAM/scripts/stlHull.x
Geometry Diagnostics¶
Track geometry analysis¶
Use the script trackAnalyzer.x to extract some information about single track from output geometry.vdb file.
Simulation parameters that are needed to extract length scale for the use in this script, are in the metadata of the geometry.vdb file.
The script prints into stdout the following x-axis dependent parameters of the track: maximal track depth and height, depth and height at the center, maximal width and width at substrate level.
Usage:
/opt/KiSSAM/scripts/trackAnalyzer.x <VDB file> > <output file>
The script works correctly if scanning strategy ScanStrategy.Type was set to SingleTrack.
This strategy implies scanning along \(x\)-axis.
On the cross-sections where \(x=\) Xcoord, the following quantities are found
Key |
Description |
|---|---|
|
\(x\) axis coordinate (mm) |
|
the (signed) distance from the lowest point of the solidified metal to the surface (mm) |
|
the (signed) distance from the highest point of the solidified metal to the surface (mm) |
|
the (signed) distance from the lowest point of the solidified metal in the center of the track (fixed \(y\)) to the surface (mm) |
|
the (signed) distance from the highest point of the solidified metal in the center of the track (fixed \(y\)) to the surface (mm) |
|
maximal width of the melted metal in the center of the track (mm) |
|
the width of the melted metal in the center of the track, measured at substrate surface (mm). Inf if there is no melted metal in this position |
Analyze Surface¶
The surfaceAnalyzer.x script outputs a 3D surface as a height function of coordinates \(h(x,y)\) in the text file format.
/opt/KiSSAM/scripts/surfaceAnalyzer.x sim_geometry.vdb > groundsurf.dat
The groundsurf.dat file contains height as a \(h(x,y)\) table, and melted material depth in the same format. Here is an example
#units: millimeters
#Xcoord Ycoord SurfaceHeight MeltpoolDepthPosition
0 0 4.564 4.564
0.007 0 4.564 4.564
0.014 0 4.564 4.564
0.021 0 4.564 4.564
0.028 0 4.564 4.564
0.035 0 4.564 4.564
...
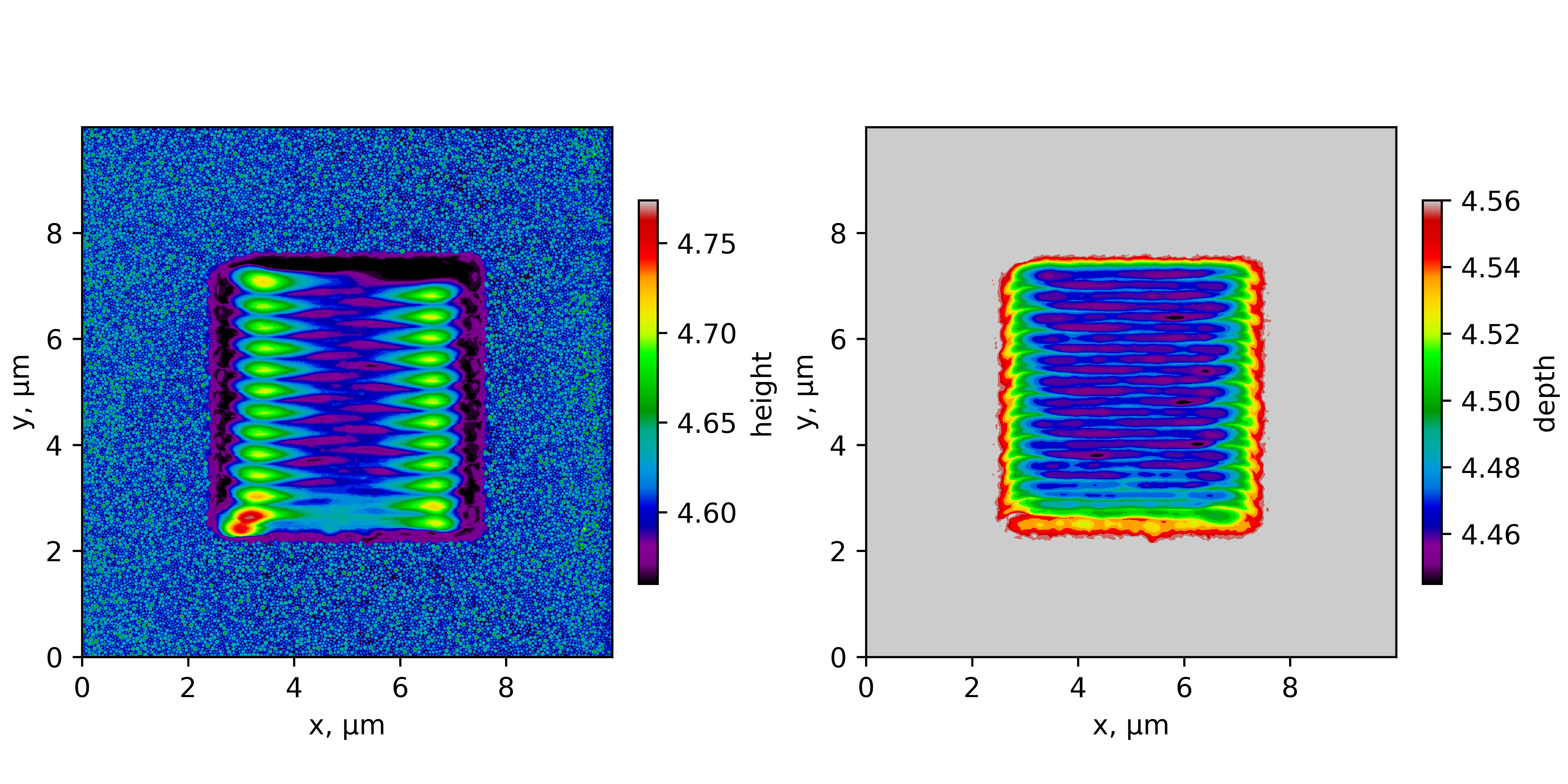
Fig. 14 Surface plot of the script output.¶
Analyze Porosity¶
The ./calcPorosity.x calculates volume and porosity map of the bulk material. Usage
/opt/KiSSAM/scripts/calcPorosity.x geometry.vdb [ --voxel_size <vx,vy,vz> ] [ --box <Xmin,Ymin,Zmin Xmax,Ymax,Zmax> ] [ --obviate_above ]
Coordinates units are millimeters. The geometry is subdivided into rectangles with the size voxel_size. The averaged porosity over each rectange is written to the output.
- --voxel_size¶
Voxel sizes are specified in mm. Voxel sizes can be set as -1. In that case, the full grid size along axis is assumed. Default voxel sizes are -1,-1,-1, and one porosity value for the whole region is computed.
- --box¶
Analyze porosity only in specified box (box sizes are specified in mm).
- --obviate_above¶
Do not consider the void region above the powder/remelted material as special region without pores.
Sample usage:
/opt/KiSSAM/scripts/calcPorosity.x drop/geometry.vdb --voxel_size 0.03,-1,-1
After the computation is finished, the output contains several columns, as in
#X-coordinate(mm) TotalVolume(mm^3) RemeltedVolume(mm^3) TotalBulkDensity,%
...
1.185 1.200390 0.000537 91.42789
1.215 1.200390 0.000539 91.48217
1.245 1.200390 0.000515 91.45513
1.275 1.200390 0.000505 91.48800
1.305 1.200390 0.000531 91.46917
1.335 1.200390 0.000528 91.46649
1.365 1.200390 0.000486 91.41384
The melted material in geometry.vdb is only the material melted in the latest layer.
In case the porosity across several layer is required, use the mergeLayers.x script in advance.
Analyze grains microstructure¶
Much information on the grains is obtained by drawing cross sections.
The following script is used to output a table with analysis of the grains.
Usage:
/opt/KiSSAM/scripts/statGrains.x geometry.vdb gtable.dat
Here, geometry.vdb is the result of KiSSAM simulation with grains growth model turned on, and
gtable.dat is a text file, where each line corresponds to one grain.
The text file contains the header where the content of the columns is explained. For each grain, there is a row which contains:
The center of mass;
Volume;
Half axes of the equivalent inertia ellipsoid (a, b, c), in the increasing order. ;
Axes of the inertia ellipsoid (3 vectors), which correspond to the a, b and c half axes;
Euler angles of the crystallographic grain orientation;
The remelted flag, which states if the grain has been remelted (None, Partially, or Fully);
The isBoundary flag, which states if the grain is at boundary of the domain, or at the boundary defined in the sizes.GrainsLimitBox parameter in the JSON file.
The script has the options:
- --only-remelted¶
Only the grains which were remelted at least partially are analyzed
- --ignore-boundary¶
будут выводиться только зерна, у которых флаг IsBoundary = No
If the statistics is needed in the portion of the domain, the geometry.vdb file can be preprocessed with the Activate Grids scripts with the –crop option.
This method can be used to output statistics of a 2D slice.